library(apa7)
library(flextable)
library(ftExtra)
library(tidyverse)
library(easystats)
library(lme4)
set_flextable_defaults(theme_fun = theme_apa,
font.family = "Times New Roman")Making tables in APA style (Part 20 of 24)
In this 24-part series, each of the tables in Chapter 7 of the Publication Manual of the American Psychological Association (7th Edition) is recreated with apa7, flextable, easystats, and tidyverse functions.
- Display of multilevel models
Figure 1
Screenshot of the APA Manual’s Table 7.20
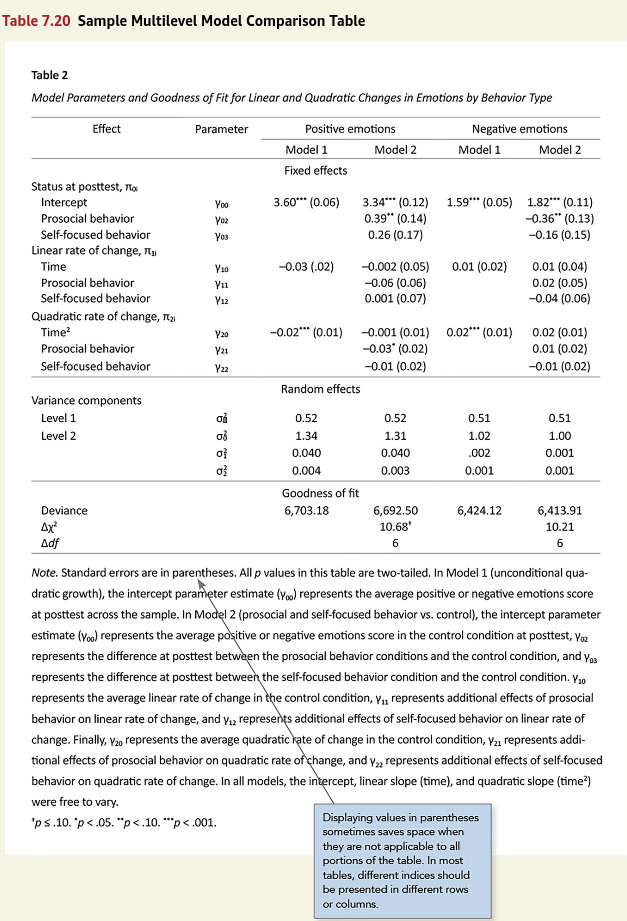
I wanted the experience of making the table close to what would happen in a real analysis. Instead of copying table’s text exactly, I simulated multilevel data based on the results in Figure 1. Although there are functions that can simulate multilevel data, In my experience, I can more easily get exactly what I want if I write out the equations and them simulate the data myself. The only information I lacked were the sample sizes at levels 1 and 2 and the number of times these behaviors were measured in the observational study. I supplied some numbers that made the models run without error.
Level 1
\begin{aligned} Y_{ij} &= b_{0j} + b_{1j}Time_{ij} + b_{2j}Time_{ij}^2 + e_{ij}\\ e_{ij}&\sim\mathcal{N}\left(0,\tau_1\right) \end{aligned}
Level 2
\begin{aligned} b_{0j} &= b_{00}+b_{01}PBS_j+b_{02}SFB_j+e_{0j}\\ b_{1j} &= b_{10}+b_{11}PBS_j+b_{12}SFB_j+e_{1j}\\ b_{2j} &= b_{20}+b_{21}PBS_j+b_{22}SFB_j+e_{2j}\\ e_2&=\begin{bmatrix} e_{0j}\\ e_{1j}\\ e_{2j} \end{bmatrix}\\ \tau_2&=\begin{bmatrix} \tau_{00}&0&0\\ 0&\tau_{11}&0\\ 0&0&\tau_{22}\\ \end{bmatrix}\\ e_2&\sim\mathcal{N}\left(0,\tau_2\right) \end{aligned}
Because of the stars after the coefficients, I was not able to find a good option for aligning both the coefficients and standard errors in the “Model” columns, so I aligned the coefficients only.
Getting coefficients, variance components, and fit statistics in the same table required a lot of custom formatting. Automating some of this would be a good idea.
set.seed(123)
# Number of people
n_2 <- 400
# Number of time points
n_1 <- 15
d_parameter <- tibble::tibble(
symbol = c(
"b_00", "b_01", "b_02",
"b_10", "b_11", "b_12",
"b_20", "b_21", "b_22",
"s_e", "s_0", "s_1", "s_2"),
Positive = c(
3.34, 0.39, 0.26, -0.002, -0.06, 0.001,
-0.001, -0.03, -0.01, 0.52, 1.31, 0.04, 0.03),
Negative = c(
1.82, -0.36, -0.16, 0.01, 0.02, -0.04, 0.02,
0.01, -0.01, 0.51, 1, 0.02, 0.02),
Parameter = c(
"(Intercept)",
"PBS",
"SFB",
"Time",
"PBS:Time",
"Time:SFB",
"I(Time^2)",
"PBS:I(Time^2)",
"I(Time^2):SFB",
"SD (Observations)",
"SD (Intercept)",
"SD (Time)",
"SD (I(Time^2))"),
pname = c(
"Intercept",
"Prosocial behavior",
"Self-focused behavior",
"Time",
"Prosocial behavior",
"Self-focused behavior",
"Time^2",
"Prosocial behavior",
"Self-focused behavior",
"Level 1",
"Level 2",
"Level 2",
"Level 2"),
l2 = c(
rep("Status at postest, π~0j~", 3),
rep("Linear rate of change, π~1j~", 3),
rep("Quadratic rate of change, π~2j~", 3),
rep("Variance components", 4)),
effect_type = c(
rep("Fixed", 9),
rep("Random", 4))
) |>
mutate(symbol = fct_inorder(symbol),
pname = fct_inorder(pname))
# Level 1 variances
tau_1 <- d_parameter |>
filter(symbol == "s_e") |>
select(symbol, Positive, Negative) |>
pivot_longer(-symbol) |>
select(name, value) |>
deframe()
# Level 2 variances
tau_2 <- d_parameter |>
filter(str_starts(symbol, "s_")) |>
filter(symbol != "s_e") |>
select(symbol, Positive, Negative) |>
pivot_longer(-symbol) |>
select(-symbol) |>
nest(value = -name) |>
mutate(value = map(
value,
\(x) diag(unlist(x)))) |>
deframe()
# Fixed effects
d_parameter_fixed <- d_parameter |>
filter(effect_type == "Fixed") |>
select(symbol, Positive, Negative) |>
pivot_longer(-symbol,
names_to = "dv",
values_to = "b") |>
pivot_wider(names_from = symbol,
values_from = b)
# Level 2 simulated data
d_2 <- tibble(
id = seq_len(n_2),
Intercept = 1,
PBS = rnorm(n_2),
SFB = rnorm(n_2)
) |>
crossing(
dv = fct_inorder(c("Positive",
"Negative"))) |>
nest(data = -dv) |>
mutate(
tau_2 = tau_2,
e_2 = map(
tau_2,
\(x) {
mvtnorm::rmvnorm(
n = n_2,
mean = c(
e_0j = 0,
e_1j = 0,
e_2j = 0),
sigma = x) |>
as_tibble()
})) |>
select(-tau_2) |>
unnest(c(data, e_2)) |>
left_join(d_parameter_fixed, by = join_by(dv)) |>
mutate(b_0j = b_00 + b_01 * PBS + b_02 * SFB + e_0j,
b_1j = b_10 + b_11 * PBS + b_12 * SFB + e_1j,
b_2j = b_20 + b_21 * PBS + b_22 * SFB + e_2j)
# Level 1 simulated data
d_1 <- d_2 |>
crossing(
Time = seq(0, n_1 - 1)
) |>
nest(data = -dv) |>
mutate(tau_1 = tau_1,
e_ij = map2(tau_1, data, \(tau, d) {
rnorm(nrow(d), sd = sqrt(tau))
})) |>
unnest(c(data, e_ij)) |>
mutate(
y = b_0j + b_1j * Time + b_2j * Time^2 + e_ij) |>
select(dv, id, PBS, SFB, Time, y)
# Analyze data
fit <- d_1 |>
nest(data = -dv) |>
mutate(
`Model 1` = map(
data,
\(d) {
lmer(y ~ 1 + Time + I(Time^2) +
(1 + Time + I(Time^2) || id),
data = d)
}),
`Model 2` = map(
data,
\(d) {
lmer(y ~ 1 + PBS * Time + PBS * I(Time^2) +
SFB * Time + SFB * I(Time^2) +
(1 + Time + I(Time^2) || id),
data = d)
})
) |>
select(-data) |>
pivot_longer(-c(dv),
names_to = "Model",
values_to = "fit") |>
mutate(
parameters = map(
fit, parameters::parameters,
include_sigma = TRUE))
# Peformance statistics
d_performance <- fit |>
select(dv, fit) |>
summarise(fit = list(fit), .by = dv) |>
mutate(p = map(fit, \(f1) {
anova(f1[[1]], f1[[2]]) |>
as_tibble() |>
rename(Deviance = `-2*log(L)`,
deltachi2 = Chisq,
df_diff = Df,
p = `Pr(>Chisq)`) |>
mutate(Model = paste0("Model ", 1:2),
chistar = p2stars(p),
Deviance = align_chr(Deviance)) |>
select(
Model,
Deviance,
deltachi2,
df_diff,
chistar) |>
apa_format_columns() |>
unite(`Δχ^2^`,
`Δχ^2^`,
chistar,
sep = "")
})) |>
select(-fit) |>
arrange(desc(dv)) |>
unnest(p) |>
mutate(dv = paste0(dv, " emotions")) |>
unite(dv, c(dv, Model)) |>
mutate(dv = fct_inorder(dv)) |>
pivot_longer(-dv, names_to = "Effect") |>
pivot_wider(names_from = dv) |>
mutate(effect_type = "Goodness of fit") |>
suppressMessages()
# Format data
d <- fit |>
unnest(parameters) |>
select(dv, Model, Parameter, Coefficient, SE, p) |>
mutate(Coefficient = align_chr(Coefficient)) |>
add_star_column(Coefficient, merge = TRUE) |>
mutate(SE = align_chr(SE,
accuracy = .01) |>
tagger(" (", ")"),
b = paste0(Coefficient, SE),
dv = fct_inorder(paste(dv, "emotions"))) |>
select(dv, Model, Parameter, b) |>
unite(dv, c(dv, Model)) |>
pivot_wider(names_from = dv, values_from = b) |>
left_join(
d_parameter |>
select(Parameter,
symbol,
pname,
l2,
effect_type),
by = join_by(Parameter)) |>
arrange(symbol) |>
rename(Effect = pname) |>
select(-Parameter) |>
relocate(Effect, symbol, .before = 0) |>
mutate(symbol = as.character(symbol) |>
str_replace_all(
c(`s_` = "σ~",
`b_` = "γ~")) |>
paste0("~", ifelse(
str_detect(symbol, "^s"),
"^2^",
""))) |>
mutate(Effect = as.character(Effect) |> str_replace("\\^2", "^2^")) |>
as_grouped_data(c("effect_type", "l2")) |>
mutate(Effect = ifelse(is.na(l2), Effect, l2)) |>
select(-l2) |>
tidyr::fill(effect_type) |>
filter(!is.na(Effect)) |>
bind_rows(d_performance) |>
mutate(across(
everything(),
\(x) replace_na(x, replace = ""))) |>
rename(`Parameter ` = symbol)
# Make table
d |>
apa_flextable(
row_title_column = effect_type,
row_title_align = "center",
font_size = 10,
line_spacing = 1.5) |>
align(j = 1, i = ~is.na(row_title)) |>
padding(
j = 1,
i = ~(is.na(row_title) &
`Parameter ` != ""),
padding.left = 15) |>
merge_v(j = "Effect") |>
width(width = c(1.70, .55,
1.05, 1.05, .05,
1.05, 1.05))Effect | Parameter | Negative emotions | Positive emotions | |||
|---|---|---|---|---|---|---|
Model 1 | Model 2 | Model 1 | Model 2 | |||
Fixed | ||||||
Status at postest, π0j | ||||||
Intercept | γ00 | 1.79*** (0.06) | 1.80*** (0.06) | 3.40*** (0.06) | 3.39*** (0.06) | |
Prosocial behavior | γ01 | −0.34*** (0.06) | 0.25*** (0.06) | |||
Self-focused behavior | γ02 | −0.17** (0.06) | 0.22*** (0.06) | |||
Linear rate of change, π1j | ||||||
Time | γ10 | 0.00 (0.01) | 0.00 (0.01) | 0.00 (0.01) | 0.00 (0.01) | |
Prosocial behavior | γ11 | 0.02 (0.01) | −0.08*** (0.01) | |||
Self-focused behavior | γ12 | −0.04*** (0.01) | 0.02 (0.01) | |||
Quadratic rate of change, π2j | ||||||
Time2 | γ20 | 0.02** (0.01) | 0.02** (0.01) | 0.02 (0.01) | 0.02 (0.01) | |
Prosocial behavior | γ21 | 0.00 (0.01) | −0.03*** (0.01) | |||
Self-focused behavior | γ22 | 0.00 (0.01) | −0.02* (0.01) | |||
Random | ||||||
Variance components | ||||||
Level 1 | σe2 | 0.73 | 0.73 | 0.71 | 0.71 | |
Level 2 | σ02 | 1.08 | 1.01 | 1.15 | 1.11 | |
σ12 | 0.13 | 0.13 | 0.22 | 0.20 | ||
σ22 | 0.13 | 0.13 | 0.18 | 0.18 | ||
Goodness of fit | ||||||
Deviance | 17,862.07 | 17,800.95 | 18,214.81 | 18,133.36 | ||
Δχ2 | 61.12*** | 81.45*** | ||||
Δdf | 6 | 6 | ||||
Table 1
Model Parameters and Goodness of Fit for Linear and Quadratic Changes in Emotions by Behavior Type
Citation
@misc{schneider2025,
author = {Schneider, W. Joel},
title = {Recreating {APA} {Manual} {Table} 7.20 in {R} with Apa7},
date = {2025-09-30},
url = {https://wjschne.github.io/posts/apatables/apa720.html},
langid = {en}
}